Scientists Apply Raman Quantitative 3D Imaging to Microbial Monitoring
Microorganisms are important contributors to the deep-sea sulfur cycle, however, the in situ detection of deep-sea microorganisms faces great challenges due to the extreme complexity of the deep-sea environment, the difficulty of sampling, isolation and cultivation of microorganisms, and the lack of near real-time nondestructive monitoring methods for microbial sulfur metabolism.
Recently, the research team led by Prof. ZHANG Xin & Prof. SUN Chaomin from the Institute of Oceanology of the Chinese Academy of Sciences (IOCAS) achieved long-term, near real-time, non-destructive microbial monitoring through 3D quantitative imaging based on the confocal Raman technology.
The study was published in mBio on Feb. 21.
In previous work, Prof. ZHANG's team found that the cold seep environment in the South China Sea was rich in elemental sulfur. Subsequently, Prof. SUN's team discovered that the cold seep bacterium Erythrobacter flavus (E. flavus) 21-3 could efficiently oxidize thiosulfate to generate elemental sulfur, and Prof. ZHANG's team identified the elemental sulfur structure as cyclic S8 by Raman spectroscopy. Then, they cooperated to in situ cultivate E. flavus 21-3 in the deep-sea cold seep and verified that the strain 21-3 could also form elemental sulfur in this environment.
Currently, the process of elemental sulfur production is mainly studied by classical biological and chemical methods, such as X-ray near-edge absorption spectroscopy, high-performance liquid chromatography, transmission electron microscopy, ion chromatography and chemometrics. However, these methods are mainly used to investigate the metabolism of microorganisms at specific time points through sampling, and cannot continuously monitor their metabolic processes on time scales without destroying the cells. Moreover, some of these methods have complicated sample preparation, which will destroy the in-situ conditions of cells. They may also result in uneven sampling and contamination, making it difficult to achieve continuous in situ observation. Therefore, new methods are urgently needed to break this bottleneck.
The confocal Raman 3D imaging has the advantages of being low-cost, rapid, label-free and non-destructive, and has the potential to perfectly combine qualitative, quantitative and visualization. To demonstrate the potential of this technique, Prof. ZHANG's team constructed Raman 3D quantitative in situ analysis method for microbial communities on solid substrates. It combined optical visualization with Raman quantitative analysis, and can quantitatively characterize the microbial metabolic processes in both temporal and spatial dimensions non-destructively. The technique has been successfully applied to the in situ monitoring of the sulfur metabolic processes of the deep-sea cold-seep bacterium E. flavus 21-3. Volume calculations and ratio analysis based on Raman 3D imaging have quantified the growth and metabolism of the microbial colony in different environments. It uncovered the unknown details of microbial growth and metabolism, and provided an important technical support for clarifying the causes of the widely distributed elemental sulfur in deep-sea cold seep.
"To our knowledge, this is the first in situ nondestructive technique to attempt long-term monitoring of microbial growth and metabolism in solid medium. It supports to rapidly identify metabolites, infer biological pathways, screen the optimal metabolic conditions of microorganisms, and compare the elemental sulfur production rate of different strains," said Prof. ZHANG.
"The successful application of this technology not only demonstrates the potential of the method for future visualization and quantitative analysis of microbial processes in situ, but also provides new ideas for studying microorganisms attached to solid surfaces in the natural environment," added Prof. SUN.
The study was supported by the National Natural Science Foundation of China, the Strategic Priority Research Program of Chinese Academy of Sciences, the Key project of Ocean Research Center and the Young Taishan Young Scholars Program.
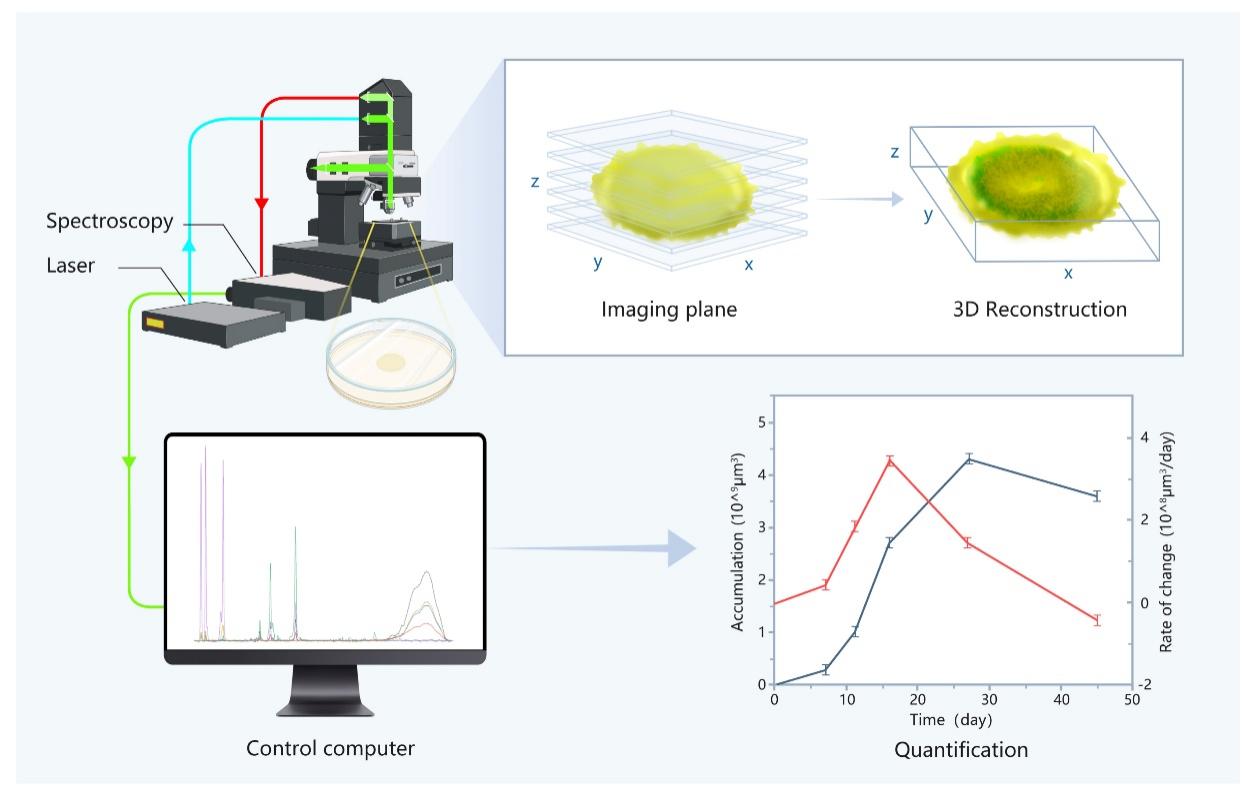
Schematic diagram of 3D quantitative imaging of colonies on solid medium
Wanying He; Ruining Cai; Shichuan Xi; Ziyu Yin; Zengfeng Du; Zhendong Luan; Chaomin Sun*, Xin Zhang*. (2023). Study of microbial sulfur metabolism in a near real-time pathway through confocal Raman quantitative 3D imaging. Microbiology Spectrum, DOI: 10.1128/spectrum.03678-22.
(Text by HE Wanying)
Media Contact:
ZHANG Yiyi
Institute of Oceanology
E-mail: zhangyiyi@qdio.ac.cn
(Editor: ZHANG Yiyi)