Excessive soil salinization has become a serious environmental and agronomic issues globally as well as in China. It is a major issues in biological science to improve the ability of plants to resist saline stress. One of the most widespread mutualistic plant-microbe relationships is that established with arbuscular mycorrhizal fungi (AMF). Nearly ninety percent of terrestrial plants, including most leguminous plants, are able to establish this type of symbiosis relationships (AM, Arbuscular mycorrhizas) with fungi of the division Glomeromycota. It is well know that AM plant had improved salt-tolerance. This beneficial effect has prompted AMF-plant associations to become a potentially useful mean in saline soil improvement. Nowadays, mechanisms underlying AM plant’s salt-tolerance is remain elusive, in spite of regulatory mechanism of arbuscular mycorrhizal formation has been well studied.
The team of “Agricultural microorganism and its application in coastal zone” in Yantai Institute of Coastal Zone Research recently discovered the salt tolerance of Sesbania cannabina plant can be enhance by Funneliformis mosseae. On this basis, signaling molecules including strigolactone and abscisic acid which may participate in the regulating AM sesbania plant’s salt-tolerance, has been identified using pharmacological and biochemical physiology means. Meanwhile, there were significant differences in gene expression patterns between AM and NM (Non-AM) plant under same saline condition. Further on, a unique gene expression groups which responded to salt stress specifically in AM plant was identified, using high throughput sequencing and weighted gene co-expression network analysis, WGCNA. Interestingly, this salt-responded gene group was exclusively expressed in shoots rather than roots which closely associated with AMF. In the same work, we also preliminarily identified that WRKY, MYB, ERF and TCP are the main regulatory transcriptional factor that participate in AM plant’s salt-tolerance. The finding will lay a molecular foundation for further understanding the regulation mechanism of AM plant’s salt-tolerance.
The relevant research results were published on the Journal of Plant Growth regulation, BMC Plant Biology and Scientific Reports, respectively. The above research works were funded by the National Natural Science Foundation of China (31601238), NSFC-Shandong Joint Fund Key Projects (U1806206) and Shandong Key Research and Development Program (2019GSF109104).
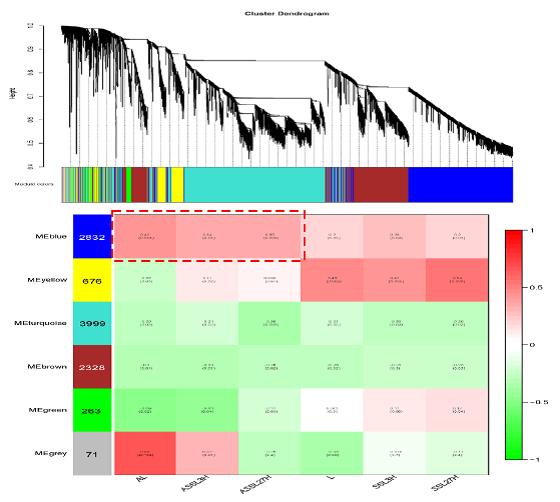
Fig.1 Module-sample association. Special gene groups related to salt stress in AM sesbania showed in red dot box. Each row corresponds to a module. The number of genes in each module is indicated on the left. Each column corresponds to a specific tissue and treatment. Each cell contains the corresponding correlation and p-value. The table is color-coded by correlation according to the color legend.
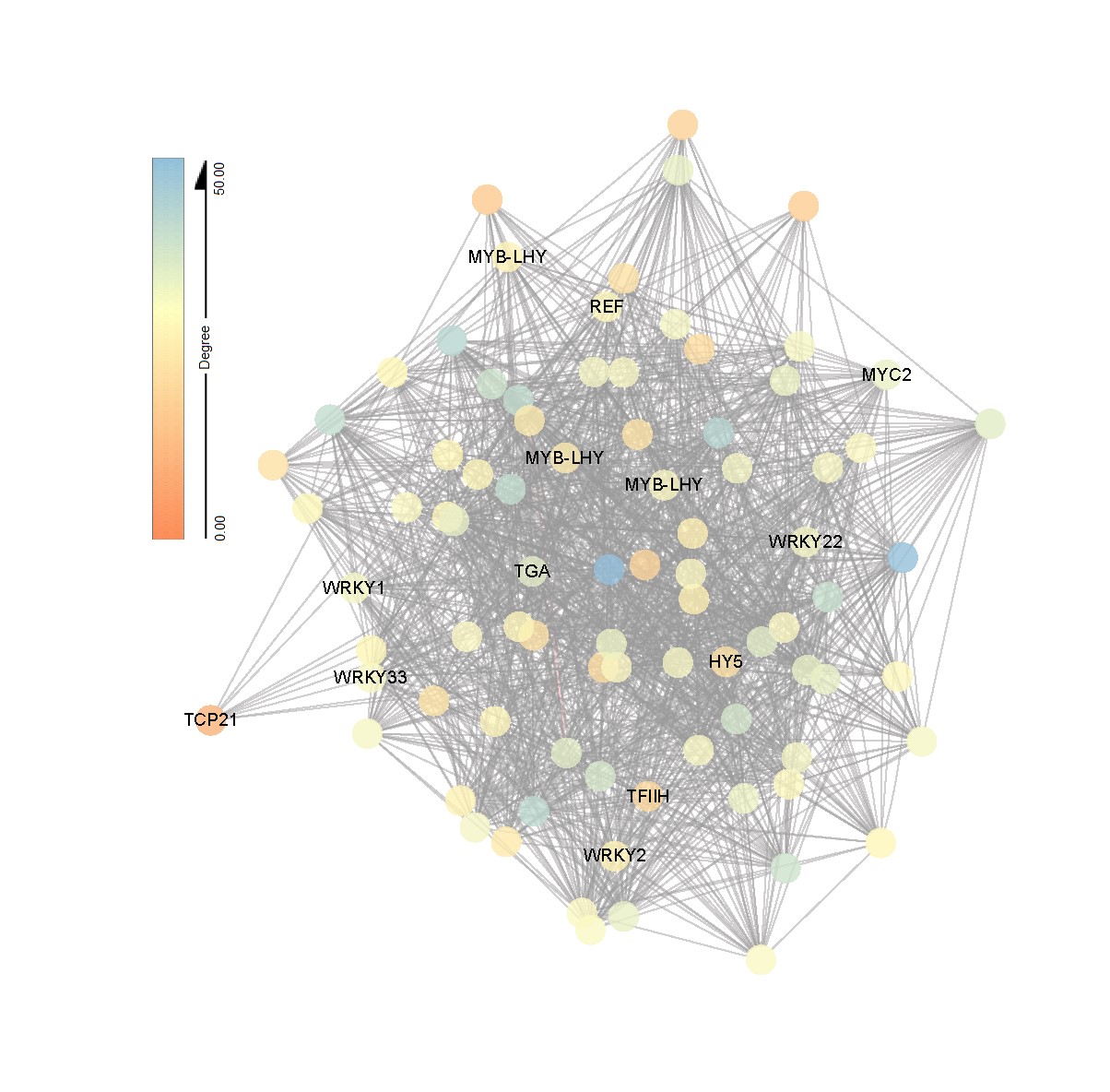
Fig.2 Transcription factors plays a key regulatory role in AM sesbania under saline stress. Network visualization of interactions between hub genes in bule module. Transcription factors are labeled with name. Eighty-five genes (top 3% of hub) are visualized by Cytoscape.