Decoding the First Shrimp Genome: A Resource of Functional Genomics and Genome Breeding for Penaeid Shrimp
Penaeid shrimp is regarded as the representative species of decapod crustaceans. Crustacean is a dominant and diverse group of aquatic animals, among which the decapods comprise many important aquatic species with high economic values, such as shrimp, crabs, lobsters, and crayfish. The Pacific white shrimp Litopenaeus vannamei is a world-wide aquaculture species with its production accounts for more than 2/3 of shrimp aquaculture production, whose output is the highest among all aquaculture species in the world. However, with the deterioration of the environment during aquaculture, disease problem and germplasm degradation are becoming the most severe threat to the shrimp aquaculture. Understanding their genetic basis to manage the pathogen infection and the genetic improvement is very urgent for shrimp species. Whole genome sequencing of shrimp will help to pave the way for solving these problems.
In fact, as early as 1997, scientists from USA proposed an international genome mapping plans for the aquatic animals, five species including salmon, catfish, tilapia, oyster and shrimp were taken as the target species based on their position in aquaculture. Till present, the genomes of four species including oyster, salmon, tilapia, catfish have been finished in succession by next generation sequencing techniques,however,the whole genome sequencing of shrimp faced a huge challenge due to their big genome size, high repeat sequence and high heterozygosity.
A research team led by Prof. Jianhai Xiang and Prof. Fuhua Li from Institute of Oceanology, Chinese Academy of Sciences (IOCAS), has successfully finished the decoding of the highly complex genome of L. under collaboration with several national and international groups. To solve above mentioned technological issues, integrated omics studies, including combination approach of the second and the third generation sequences, transcriptomics, proteomics and bioinformatics, have been implemented. Finally, the first shrimp de novo reference genome in the world has been completed through efforts of more than 10 years. This research progress was published in the top journal Nature Communications on January 21, 2019 with the title "Penaeid shrimp genome provides insights into benthic adaptation and frequent molting".
The L. vannamei genome covers ~1.66 Gb (scaffold N50 605.56 Kb) with 25,596 protein-coding genes. It holds the highest proportion of simple sequence repeats (SSRs, more than 23.93%) among the sequenced animals, presenting a challenge for genome sequencing and assembly. The high quality genome assembly of L. vannamei provides the opportunity to understand various biological processes of shrimp at the genome level. The most prominent characteristic of the coding region of the shrimp genome is the expansion of a series of genes related to visual system, nerve signal conduction and molting. These gene expansions might have provided the shrimp with its excellent eyesight and rapid nerve signal conduction, better equipping it to adapt to its benthic habitat. The shrimp genome assembly also sheds light on the molting process and provides important evidences for exploring the similarities and differences between crustaceans and other ecdysozoans, including insects. As a vital process in shrimps, molting is tightly linked to growth, metamorphosis and reproduction. Elucidating the regulatory mechanisms of molting will provide useful clues for genetic analysis of these important processes. The genomic analysis on the breeding population and wild population of shrimp reveals that the farmer’sbreeding practice of only 30 years has already exerted a significant impact on the genome of the L. vannamei broodstocks. An increase in aquaculture production and efficiency can be achieved through genetic improvement of cultured stocks. The assembled shrimp genome and the large amount of SNP markers will provide a useful resource for the application of genome-wide association studies and genomic selection, and thus accelerate genetic improvements in shrimp culture.
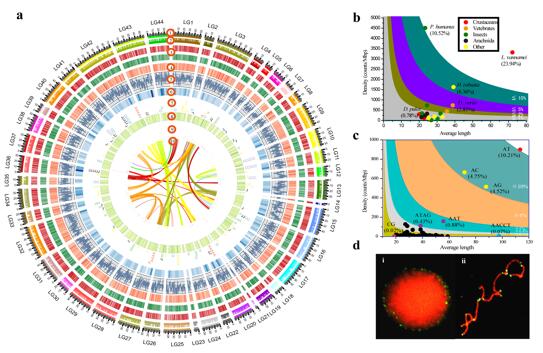
The genome map of Litopenaeus vannamei and the characteristic of simple sequence repeats (SSRs)
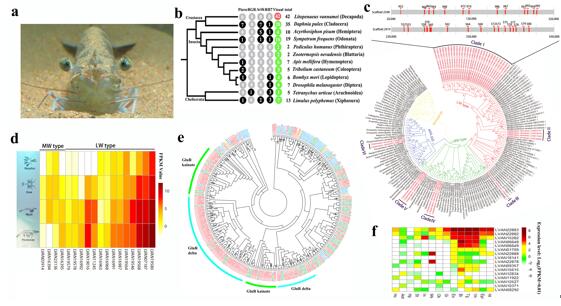
The Opsin and iGluR gene families in the shrimp genome to show its adaptions to benthic environment.
Reference:Zhang X, Yuan J, Sun Y, Li S, Gao Y, Yu Y, Liu C, Wang Q, Lv X, Zhang X, Ma KY, Wang X, Lin W, Wang L, Zhu X, Zhang C, Zhang J, Jin S, Yu K, Kong J, Xu P, Chen J, Zhang H-B, Sorgeloos P, Sagi A, Alcivar-Warren A, Liu Z, Wang L, Ruan J, Chu KH, Liu B, Li F, Xiang J. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat Commun. (2019) 10: 356. https://doi.org/10.1038/s41467-018-08197-4